
The Centre for Enzyme Innovation
At the Centre for Enzyme Innovation, we are working to solve one of the most pressing environmental issues facing our planet.
Learning from the natural world, we are delivering transformative enzyme-enabled solutions for the circular recycling of plastics.
Following a £5.8 million award from the Research England Expanding Excellence Fund in 2019, and a £1m award from the Solent Local Enterprise Partnership in 2020 through the HM Government Getting Building Fund, we have created state-of-the-art facilities and built a team of specialist multidiscipline researchers to deliver world-class research and innovation.
Our research
We currently have 30 scientists covering a wide range of disciplines including microbiology, molecular biophysics, biochemistry, enzyme engineering and synthetic biology, biotechnology, and more recently, polymer chemistry.
Hosted in our new custom laboratories, we have the expertise and facilities required to help tackle the challenge of plastic pollution and develop enzyme-based low energy, low carbon, biorecycling solutions.
Our research sits at the interface between enzymes and polymers with a focus on both pure and applied research in biocatalysis.
We are expanding our research and innovation activities to address the diverse range of plastics, including mixed waste streams and composites, materials that are often incinerated or end up in landfill and leak to the environment.
Centre Director
Our research is divided into 4 areas
- Discover new enzymes from the environment that break down plastics
- Engineer these enzymes to enhance their activity, stability and yield
- Deploy enzymes by pilot scale fermentation and industry-ready formulations
- Apply these enzymes in proof-of-concept biorecycling and upcycling processes
How we work
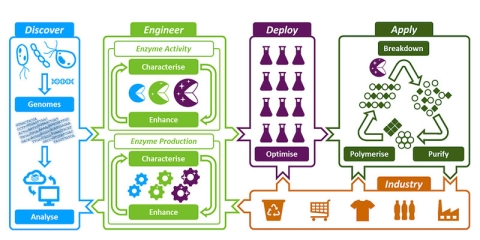
The CEI’s unique integrated research and innovation pipeline
The pipeline allows for the streamlined development of enzymes from discovery through to recycling applications.
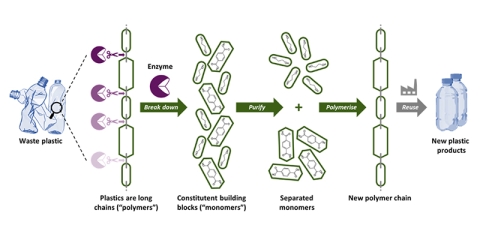
Working towards a circular plastics economy
Enzymes break down waste plastic polymers into building blocks that are purified and re-polymerised, allowing infinite recycling of the materials as part of a circular plastics economy.
Recent research outputs
Investigating the effect of fusion partners on the enzymatic activity and thermodynamic stability of poly(ethylene terephthalate) degrading enzymes
Oliveira Pessoa, L., Cahill, A., Wuscher, L., Green, K.R., Bemmer, V.L., Lichtenstein, B.R., 2024, Faraday Discussions, early-online d4fd00067f
Plastics pose significant environmental challenges due to their durability. Engineered enzymes like PETases offer a sustainable recycling solution but struggle with plastic's hydrophobic surface, product inhibition, and absence of processivity. Fusion partners have been used to address these obstacles, however there is ambiguity in whether fusion proteins provide a tangible benefit. This study uses SpyCatcher003, a thermodynamically switchable fusion partner, to investigate its effect on PETase activity and stability. Our results indicate that for small, reversibly folding fusion partners, enzyme stability and tolerance to fusions is more important than fusion partner properties. This approach should prove generalisable in isolating and clarifying the thermodynamic benefits of fusion partners on enzymes for biotechnological applications.
Concentration-dependent inhibition of mesophilic PETases on poly(ethylene terephthalate) can be eliminated by enzyme engineering
Avilan, L., Lichtenstein, B., Koenig, G., Zahn, M., Allen, M.D., Oliveira Pessoa, L., Clark, M.A., Bemmer, V.L., Graham, R., Austin, H.P., Dominick G., Johnson, C.W., Beckham, G.T., McGeehan, J., and Pickford, A., 2023 (Early online) ChemSusChem.
Enzyme-based depolymerization, particularly PETase from Ideonella sakaiensis, offers a promising method for poly(ethylene terephthalate) (PET) recycling, despite its concentration-dependent inhibition which varies depending on factors like incubation time, solution conditions, and PET surface area. This inhibitory trait is also observable in other mesophilic PET-degrading enzymes, regardless of their PET depolymerization activity level, and is not tied to any obvious structural basis. Significantly, thermostable PETase variants, including the engineered HotPETase, show less or no inhibition, pointing to the benefit of enhanced thermostability in these enzymes for PET hydrolysis.
Initiation of fatty acid biosynthesis in Pseudomonas putida KT2440
McNaught, K., Kuatsjah, E., Zahn, M., Prates, E., Shao, H., Bentley, G., Pickford, A., Gruber, J., Hestmark, K., Jacobson, D., Poirier, B., Ling, C., San Marchi, M., Michener, W.E., Nicora, C., Sanders, J., Szostkiewicz, C.J., Veličković, D., Zhou, M., Monoz, N., and 7 others, 2023, Metabolic Engineering 76, p. 193-203.
Understanding bacterial fatty acid biosynthesis is vital for producing fatty acid-derived molecules and developing antibiotics, but knowledge gaps persist regarding its initiation. Our study shows that the microbe Pseudomonas putida KT2440 initiates fatty acid biosynthesis through three distinct pathways involving enzymes FabH1, FabH2, and MadB. Through comprehensive in vivo and in vitro testing, along with X-ray crystallography and computational modelling, we have identified the possible mechanism of malonyl-ACP decarboxylation via MadB, a pathway common in bacteria that offers potential for biotechnological and medical applications.
Biochemical and structural characterization of a Sphingomonas diarylpropane lyase for cofactorless deformylation
Kuatsjah, E., Zahn, M., Chen, X., Kato, R., Hinchen, D.J., Konev, M., Katahira, R., Orr, C., Wagner, A., Zou, Y., Haugen, S.J., Ramirez, K., Michener, J.K., Pickford, A., Kamimura, N., Masai, E., Houk, K.N., McGeehan, J. and Beckham, G.T., 2023, Proceedings of the National Academy of Sciences of the United States of America, 120, 4, 9 p., e2212246120.
Efforts to valorize lignin through catalytic depolymerization and biological funneling require the development of enzymes that can break down aromatic dimers and oligomers to monomers. Our study examines LdpA, an enzyme found in Novosphingobium aromaticivorans and Sphingobium sp. SYK-6, that transforms a lignin-derived dimer, erythro-1,2-diguaiacylpropane-1,3-diol (erythro-DGPD), into lignostilbene, demonstrating this transformation lacks enantioselectivity and is impeded by product inhibition. Using X-ray crystallography, we identified key catalytic residues and extended the scope of the NTF-2-like protein family's chemistry to include lignin dimer deformylation, a reaction not requiring a cofactor.
Microbiome species diversity and seasonal stability of two temperate marine sponges Hymeniacidon perlevis and Suberites massa
Lamb, C.E. and Watts, J.E.M. 2023, Environmental Microbiome 18, 1, 11 p., 52.
Marine sponges possess diverse microbiomes that have shown changes in response to environmental factors such as nutrient availability, temperature, and light. This study investigates the impact of temperature shifts on the composition and function of the microbiome in two UK-native marine sponge species, using next-generation sequencing. The research identifies a stable, species-specific core microbiome unaffected by seasonal temperature variations, potentially enabling the discovery of new functional activities in the marine ecosystem.
Identification of small RNAs associated with RNA chaperone Hfq reveals a new stress response regulator in Actinobacillus pleuropneumoniae
da Silva, G.C., Rossi, C.C., Rosa, J.N., Sanches, N.M., Cardosa, D.L., Li, Y., Witney, A.A., Gould, K.A., Fontes, P.P., Callaghan, A.J., Bossé, J.T., Langford, P.R., and Bazzolli, D.M.S., 2022, Frontiers in Microbilogy 13, 20 p., 1017278
The RNA chaperone Hfq and its associated small RNAs (sRNAs) are known to influence the virulence and stress response of Actinobacillus pleuropneumoniae. This study identified 14 sRNAs linked with Hfq in A. pleuropneumoniae strain MIDG2331, focusing on Rna01, an sRNA with potential stress regulon recognition, which affects biofilm formation, virulence, stress susceptibility, and outer membrane protein composition. This research advances our understanding of Hfq-dependent sRNAs in bacterial physiology and pathogenicity.
Versatile and facile one-pot biosynthesis for amides and carboxylic acids in E. coli by engineering auxin pathways of plant microbiomes
Menon, N., Richmond, D., Rahman, M.R. and Menon, B.R.K., 2022, ACS Catalysis 12, 4, p.2309-2319
Creating biocatalytic methods for incorporating amide or carboxylic acid functional groups into bioactive scaffolds in aqueous conditions poses a significant challenge due to the dearth of suitable enzymes. This study presents a monooxygenase-hydrolase based biosynthetic method, leveraging pathways from plant microbiomes involved in auxin production, a plant hormone used for infecting and colonizing plant cells. Demonstrating the versatility of this method, we designed one-pot multienzyme biosynthetic cascades for large-scale production of indole acetic acid and its derivatives.
A review of cross-disciplinary approaches for the identification of novel industrially relevant plastic-degrading enzymes
Herbert, J., Beckett, A.H., and Robson, S.C., 2022, Sustainability 4, 23, 25p., 15898.
This review explores interdisciplinary methods for identifying environmental enzymes that can break down plastic polymers into monomeric building blocks, offering a solution to mounting plastic waste through circular recycling. Techniques such as DNA and RNA sequencing of plastic-degrading microbes, alongside machine learning, aid in pinpointing industrially viable enzymes, which can be optimized via protein engineering. These combined approaches offer promise for identifying additional enzyme candidates to tackle the global plastic issue at an industrial scale.
The role of binding modules in enzymatic poly(ethylene terephthalate) hydrolysis at high solids loadings
Graham, R., Erickson, E., Brizendine, R.K., Salvachúa, D., Michener, W.E., Li, Y., Tan, Z., Beckham, G.T., McGeehan.J., Pickford, A.R., 2022, Chem Catalysis 2, 10, p. 2644-2657.
Enzymes deconstructing biological polymers often possess multi-domain structures, including a catalytic domain and a non-catalytic binding module, which enhances enzyme concentration on the substrate surface and boosts the hydrolysis of poly(ethylene terephthalate) (PET) in engineered cutinase enzymes. In this study, we created fusion constructs of leaf compost cutinase (LCC) with carbohydrate-binding modules (CBMs), which improved aromatic monomer yield from PET at solids loadings under 10 wt%, but didn't provide additional benefits at higher loadings. This indicates that fusion constructs with these non-catalytic binding modules aren't required for industrial enzymatic PET recycling.
Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Erickson, E., Gado, J.E., Avilan, L., Bratti, F., Brizendine, R.K., Cox, P., Gill, R., Graham, R., Kim, D-J., Koenig, G., Michener, W.E., Poudel, S., Ramirez, K.J., Shakespeare, T.J., Zahn, M., Boyd, E.S., Payne, C., DuBois, J.L., Pickford, A.R., Beckham, G.T., and 1 others, 2022, Nature Communications 13, 1, 15 p., 7850.
The study uses bioinformatics and machine learning to identify 74 potential thermotolerant PET hydrolases from natural diversity, aimed at augmenting the enzymatic deconstruction of poly(ethylene terephthalate) (PET). Of these, 51 enzymes from seven phylogenetic groups were successfully produced, purified, and assayed, revealing PET hydrolysis activity across a range of pH values and temperatures, with different substrate selectivities based on PET morphology. Using X-ray crystallography and AlphaFold, the study also uncovers novel protein folds and accessory domains linked with PET deconstruction, expanding the range of thermotolerant scaffolds for this purpose.
Comparative performance of PETase as a function of reaction conditions, substrate properties, and product accumulation
Erickson, E., Shakespeare, T. J., Bratti, F., Buss, B., Graham, R., Hawkins, M., Koenig, G., Michener, W. E., Miscall, J., Ramirez, K., Rorrer, N. A., Zahn, M., Pickford, A., McGeehan, J. E. and Beckham, G. T. 2022, ChemSusChem 15, 1, 11 p., e202101932.
This study contrasts the Ideonella sakaiensis PETase wild-type enzyme with an improved variant for PET recycling, revealing that reaction temperature, substrate morphology, and reaction mixture composition significantly impact performance. It shows higher PET conversion for substrates with moderate crystallinity and distinct inhibition profiles for each enzyme, dependent on the accumulation of certain products. The findings underscore the critical role of reaction conditions, substrate selection, and product accumulation in the performance of PET-hydrolyzing enzymes, vital for enzyme screening in developing enzyme-based polyester recycling.
Funding
Researchers from the Centre for Enzyme Innovation receive funding from a wide range of external sources, including the Biotechnology and Biological Sciences Research Council (BBSRC), the National Environment Research Council (NERC), the Engineering and Physical Sciences Research Council (EPSRC), the European Commission, Innovate UK, Diamond Light Source, the Defence Science and Technology Laboratory (DSTL), the U.S. Department of Energy National Renewable Energy Laboratory (NREL), Johnson Matthey, the Royal Society, the Royal Society of Chemistry, and the Iwatani Foundation.
Our most recent large funding awards were awarded from the Research England E3 scheme (£5.8m) and the Solent Local Enterprise Partnership (£1m).





Contact us
Discover how you can collaborate with the CEI.
If you're a commercial or academic organisation interested in working with us, please contact our Industry Engagement Lead, Dr Victoria Bemmer.






